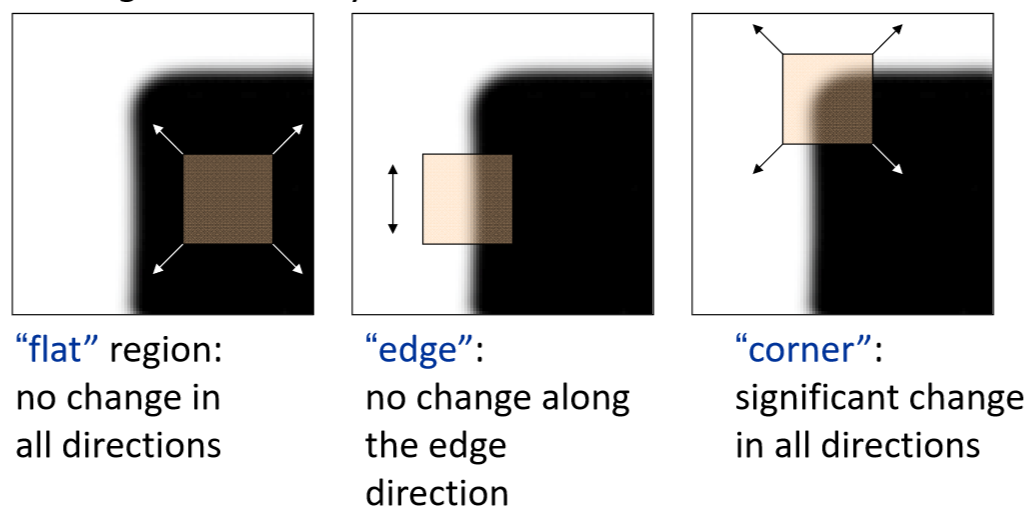
Basic idea
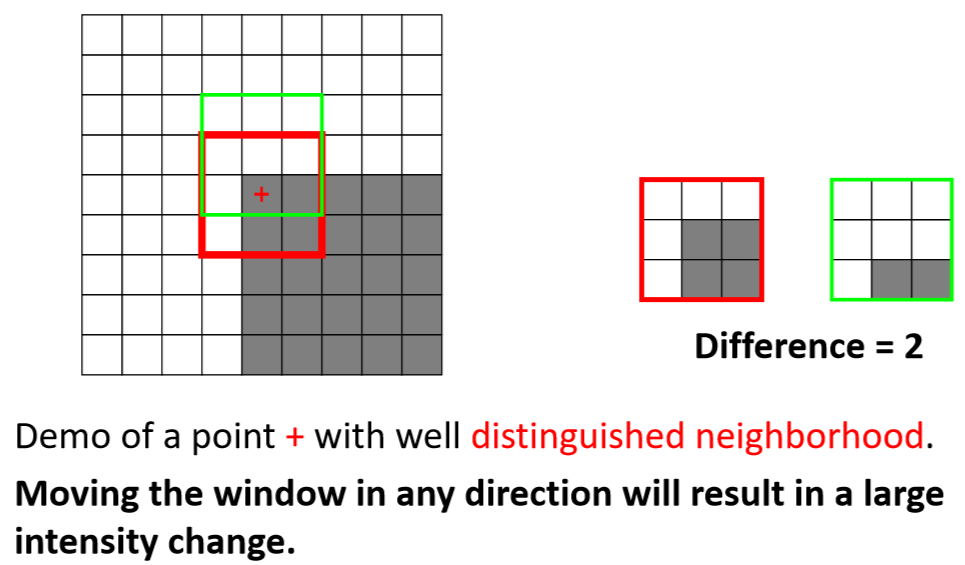
Algorithm
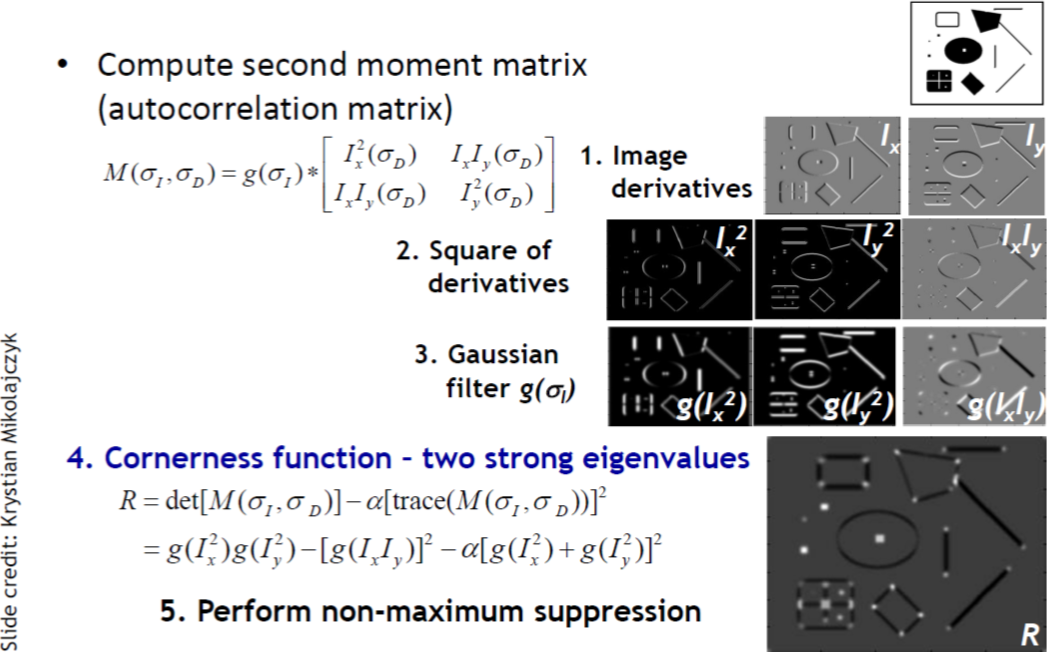
Code
main.m 主函数
im = imread('officegray.bmp');
im = double(im);
sigma = 2.5;
thresh = 35;
nonmaxrad = 5;
K=0.02;
[cim, r3, c3] = harris(im, sigma,thresh, nonmaxrad);
figure;
imshow(im,[]), hold on, plot(c3,r3,'*');
harris.m 角点检测
function [cim, r, c, rsubp, csubp] = harris(im, sigma, thresh, radius, disp)
error(nargchk(2,5,nargin));
if nargin == 4
disp = 0;
end
if ~isa(im,'double')
im = double(im);
end
subpixel = nargout == 5;
[Ix, Iy] = derivative5(im, 'x', 'y'); %Ix,Iy是
Ix2 = gaussfilt(Ix.^2, sigma);
Iy2 = gaussfilt(Iy.^2, sigma);
Ixy = gaussfilt(Ix.*Iy, sigma);
k = 0.04;
cim = (Ix2.*Iy2 - Ixy.^2) - k*(Ix2 + Iy2).^2; % 计算公式.
if nargin > 2 % We should perform nonmaximal suppression and threshold
if disp % Call nonmaxsuppts to so that image is displayed
if subpixel
[r,c,rsubp,csubp] = nonmaxsuppts(cim, radius, thresh, im);
else
[r,c] = nonmaxsuppts(cim, radius, thresh, im);
end
else % Just do the nonmaximal suppression
if subpixel
[r,c,rsubp,csubp] = nonmaxsuppts(cim, radius, thresh);
else
[r,c] = nonmaxsuppts(cim, radius, thresh);
end
end
end
derivative5.m 计算梯度
function varargout = derivative5(im, varargin)
varargin = varargin(:);
varargout = cell(size(varargin));
secondDeriv = false;
for n = 1:length(varargin)
if length(varargin{n}) > 1
secondDeriv = true;
break
end
end
if ~secondDeriv
p = [0.037659 0.249153 0.426375 0.249153 0.037659];
d1 =[0.109604 0.276691 0.000000 -0.276691 -0.109604];
else
p = [0.030320 0.249724 0.439911 0.249724 0.030320];
d1 = [0.104550 0.292315 0.000000 -0.292315 -0.104550];
d2 = [0.232905 0.002668 -0.471147 0.002668 0.232905];
end
gx = false;
for n = 1:length(varargin)
if strcmpi('x', varargin{n})
varargout{n} = conv2(p, d1, im, 'same');
gx = true; % Record that gx is available for gxy if needed
gxn = n;
elseif strcmpi('y', varargin{n})
varargout{n} = conv2(d1, p, im, 'same');
elseif strcmpi('xx', varargin{n})
varargout{n} = conv2(p, d2, im, 'same');
elseif strcmpi('yy', varargin{n})
varargout{n} = conv2(d2, p, im, 'same');
elseif strcmpi('xy', varargin{n}) | strcmpi('yx', varargin{n})
if gx
varargout{n} = conv2(d1, p, varargout{gxn}, 'same');
else
gx = conv2(p, d1, im, 'same');
varargout{n} = conv2(d1, p, gx, 'same');
end
else
error(sprintf('''%s'' is an unrecognized derivative option',varargin{n}));
end
end
Gaussfilt.m 高斯模糊
function smim = gaussfilt(im, sigma)
assert(ndims(im) == 2, 'Image must be greyscale');
if ~strcmp(class(im),'double')
im = double(im);
end
sze = ceil(6*sigma);
if ~mod(sze,2)
sze = sze+1;
end
sze = max(sze,1);
h = fspecial('gaussian', [sze sze], sigma);
smim = filter2(h, im);
nonmaxsuppts.m 非极大值抑制
function [r,c, rsubp, csubp] = nonmaxsuppts(cim, radius, thresh, im)
subPixel = nargout == 4; % We want sub-pixel locations
[rows,cols] = size(cim);
% Extract local maxima by performing a grey scale morphological
% dilation and then finding points in the corner strength image that
% match the dilated image and are also greater than the threshold.
sze = 2*radius+1; % Size of dilation mask.
mx = ordfilt2(cim,sze^2,ones(sze)); % Grey-scale dilate.
% Make mask to exclude points within radius of the image boundary.
bordermask = zeros(size(cim));
bordermask(radius+1:end-radius, radius+1:end-radius) = 1;
% Find maxima, threshold, and apply bordermask
cimmx = (cim==mx) & (cim>thresh) & bordermask;
[r,c] = find(cimmx); % Find row,col coords.
if subPixel % Compute local maxima to sub pixel accuracy
if ~isempty(r) % ...if we have some ponts to work with
ind = sub2ind(size(cim),r,c); % 1D indices of feature points
w = 1; % Width that we look out on each side of the feature
% point to fit a local parabola
% Indices of points above, below, left and right of feature point
indrminus1 = max(ind-w,1);
indrplus1 = min(ind+w,rows*cols);
indcminus1 = max(ind-w*rows,1);
indcplus1 = min(ind+w*rows,rows*cols);
% Solve for quadratic down rows
rowshift = zeros(size(ind));
cy = cim(ind);
ay = (cim(indrminus1) + cim(indrplus1))/2 - cy;
by = ay + cy - cim(indrminus1);
rowshift(ay ~= 0) = -w*by(ay ~= 0)./(2*ay(ay ~= 0)); % Maxima of quadradic
rowshift(ay == 0) = 0;
% Solve for quadratic across columns
colshift = zeros(size(ind));
cx = cim(ind);
ax = (cim(indcminus1) + cim(indcplus1))/2 - cx;
bx = ax + cx - cim(indcminus1);
colshift(ax ~= 0) = -w*bx(ax ~= 0)./(2*ax(ax ~= 0)); % Maxima of quadradic
colshift(ax == 0) = 0;
rsubp = r+rowshift; % Add subpixel corrections to original row
csubp = c+colshift; % and column coords.
else
rsubp = []; csubp = [];
end
end
if nargin==4 & ~isempty(r) % Overlay corners on supplied image.
figure(1), imshow(im,[]), hold on
if subPixel
plot(csubp,rsubp,'r+'), title('corners detected');
else
plot(c,r,'r+'), title('corners detected');
end
hold off
end
if isempty(r)
fprintf('No maxima above threshold found\n');
end
效果图